Local background correction
Prerequisites
Before starting this lesson, you should be familiar with:
Learning Objectives
After completing this lesson, learners should be able to:
Understand how to use image filters for creating a local background image
Use the generated local background image to compute a foreground image
Motivation
Very often, biological images contain locally varying background intensities. This hampers both segmentation and intensity quantification. However, given a sufficient separation of length scales in terms of variation in background intensities vs. variation in intensities in the foreground, image filters can be employed to measure and correct for the background.
Concept map
graph LR
i(Input image) --> bgf(Large size filter)
bgf --> bgi[Background image]
bgi --> s[Subtract]
i --> s
s --> fgi[Foreground image]
Example figure
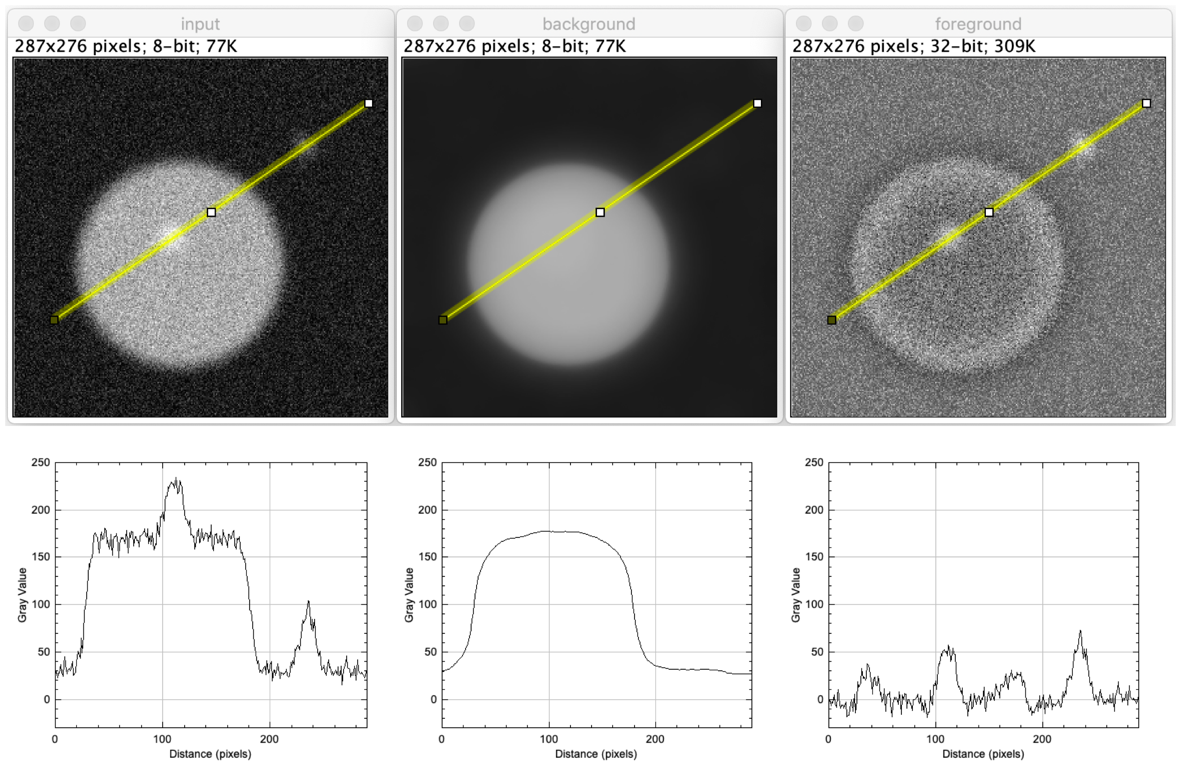
Common filters for creating background images
- Median filter
- “Rolling ball” background
- ImageJ’s “Subtract Background”
- Missing: explanation how it works, exactly
- ImageJ’s “Subtract Background”
- Morphological opening filter
- The result of background subtraction using a grayscale opening is called top-hat filter
Advanced and powerful filters for creating background images
- Morphological opening using reconstruction
- How do the work?
- MATLAB has this option…
imreconstruct - ImageJ MorpholibJ has a Grayscale Attribute Filtering, which seems similar (the same)?
Activity
- Open image xy_8bit__some_spots_with_uneven_bg
- Compute a background image using a median filter
- Create a foreground image by subtracting the background image from the input image
- Check how well the background corrrection worked and whether any artefacts were introduced
- Optional: discuss how one could automatically segment the two spots in the resulting foreground image (mean filter and object size filter)
ImageJ GUI & Macro
run("Close All"); // Open data open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit__spots_local_background_with_noise.tif"); rename("input"); // Create background image run("Duplicate...", "title=background"); run("Median...", "radius=30"); rename("background"); // Create foreground image imageCalculator("Subtract create 32-bit", "input","background"); rename("foreground"); run("Tile"); // Make line profiles to quantitatively display what happened run("Line Width...", "line=7"); selectWindow("input"); makeLine(30, 201, 272, 34); run("Plot Profile"); Plot.setLimits(0,290,-30.0,250); selectWindow("background"); makeLine(30, 201, 272, 34); run("Plot Profile"); Plot.setLimits(0,290,-30.0,250); selectWindow("foreground"); makeLine(30, 201, 272, 34); run("Plot Profile"); Plot.setLimits(0,290,-30.0,250);
Exercises
Show exercises for:ImageJ GUI & Macro
Spots in a cell
Note: This module’s activity (see above) contains all the ImageJ commands that you need for below exercise.
- Open xy_16bit__autophagosomes_crop.tif
- Duplicate the image and apply a median filter in order to create the background image
- Choose the radius of the median filter just large enough such that the bright spots dissappear
- Subtract the resulting background image from the input image
- You should see an image with the bright spots, but now without the uneven background
Solution
run("Close All"); // File › Open... open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_16bit__autophagosomes_crop.tif"); // Image › Rename... rename("input"); // Image › Duplicate... run("Duplicate...", "title=median"); // Process › Filters › Median... run("Median...", "radius=7"); // Image › Rename... rename("background"); // Process › Image Calculator... imageCalculator("Subtract create 32-bit", "input", "background"); // Image › Rename... rename("foreground"); // Window › Tile run("Tile");Motors on microtubules (advanced)
The magic of area openings….
- Open xy_16bit__motors_on_microtubules.tif
- The challenge is to automatically segment and measure the intensity of the two spots (motors)
- Try to create a background image only containing the microtubules, using a median filter
- Subtract the resulting background image from the input image
- You will probably not succeed in getting a clean result that only contains the motors.
- Why is this the case?
- Try now creating a background image using [ Plugins › MorphoLibJ › Gray Scale Attribute Filtering ]
- It works, it’s magic, isn’t it?
Solution
run("Close All"); open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_16bit__motors_on_microtubules.tif"); rename("input") run("Duplicate...", "bg"); run("Gray Scale Attribute Filtering", "operation=Opening attribute=Area minimum=100 connectivity=4"); rename("bg"); imageCalculator("Subtract create 32-bit", "input","bg"); run("Tile")
Assessment
True or false (discuss with your neighbour)
- Mean filter is better than the median filter to generate background image.
- On the generated background image the objects of interest should not be visible.
- The size of the filter’s structuring element for generating the background image should be much smaller than the size of the objects.
Solution
- False (mean filter is really quite poor in terms of removing foreground information)
- True (because this is the background image, so it should not contain any foreground information)
- False (it should be much (maybe ~3 times) larger)
Follow-up material
We recommend reading these modules next:
Learn more: