Connected component labeling
Prerequisites
Before starting this lesson, you should be familiar with:
Learning Objectives
After completing this lesson, learners should be able to:
Understand how objects in images are represented as a label mask image.
Apply connected component labeling to a binary image to create a label mask image.
Motivation
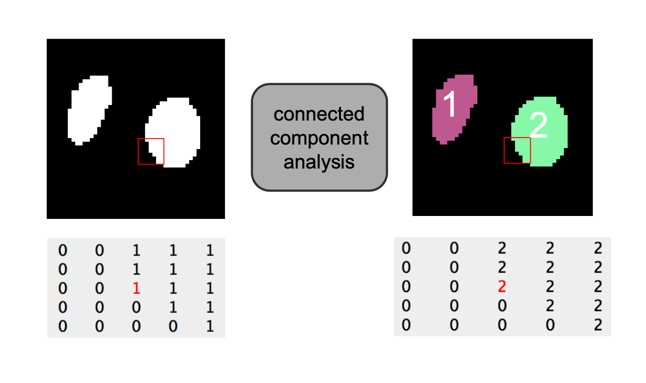
In bioimage analysis one very often wants to detect objects or specific regions in images. A typical workflow is to first categorise an image into in background and foreground regions, which can be represented as a binary image. The next step is a connected components labeling, where spatially connected regions of foreground pixels are assigned (labeled) as being part of one region (object).
Concept map
Example figure
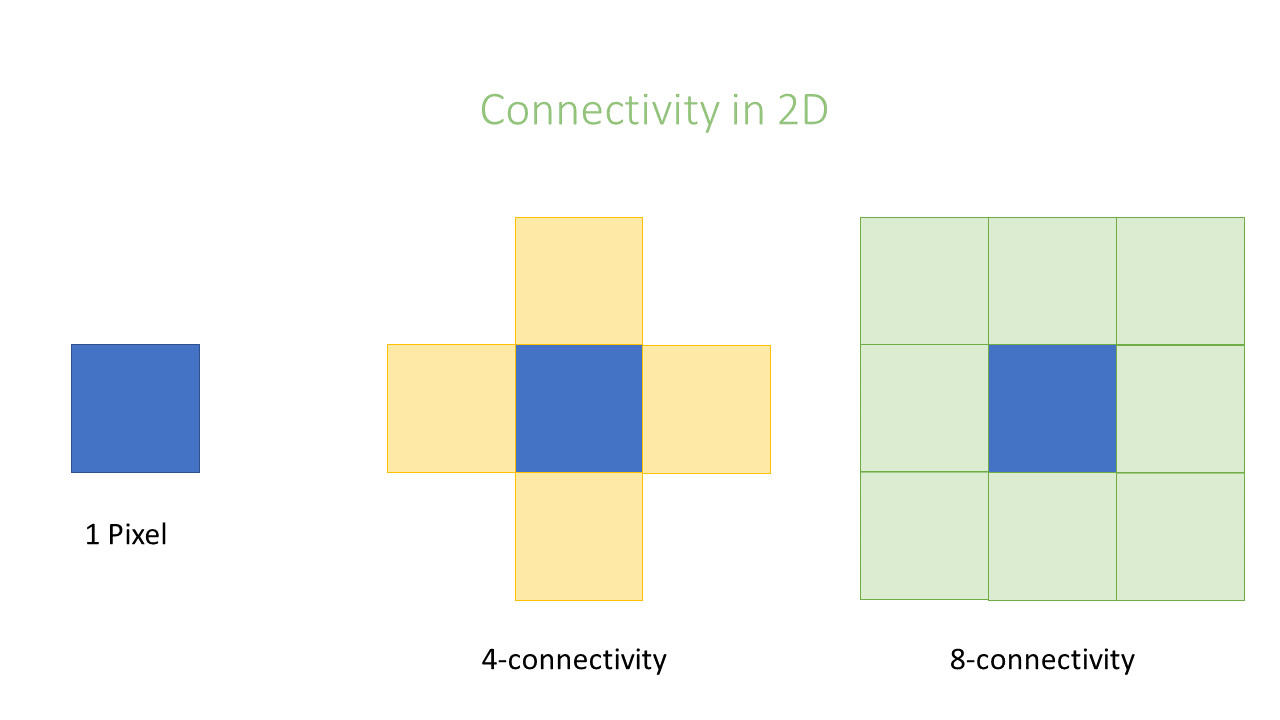
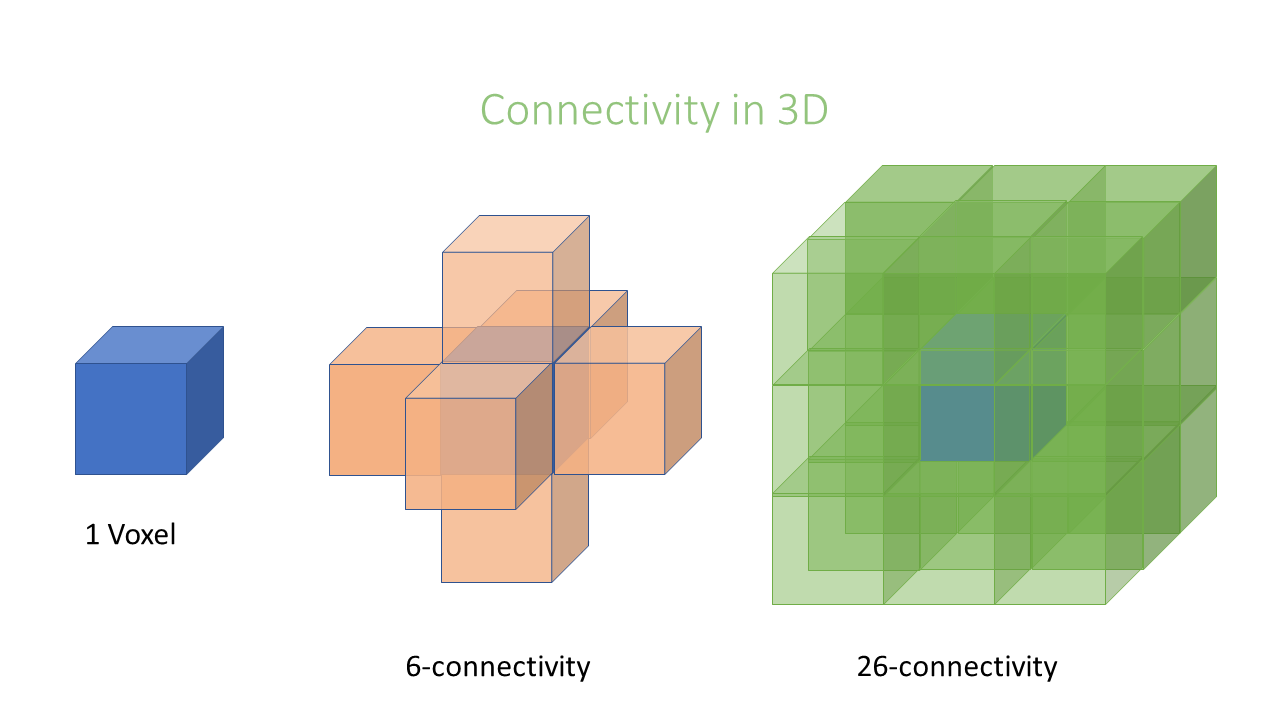
Connectivity
In an image pixels are ordered in a squared configuration. For performing a connected component analysis
it is important to define which pixels are considered direct neighbors of a pixel. This is called connectivity and defines which
pixels are considered connected to each other. Unfortunately, there is not only one way to define connectivity. Depending,
if we consider corner pixels or not we can have in 2D a 4 or 8–connectivity, respectively, in 3D a 6 or 26-connectivity. Which connectivity
value we choose can affect the object size.
Activity
- 2D connected component labeling:
- Open image xy_8bit_binary__nuclei.tif.
- Create a label image by means of 4-connected connected components labeling.
- Apply a multi-color LUT.
- Repeat with 8-connected labeling and discuss the difference.
- 3D connected component labeling:
- Open image xyz_8bit_binary__spots.tif
- Inspect the histogram of the label image and discuss what you can learn from it.
ImageJ MorpholibJ Macro & GUI
/** * 2D and 3D connected components labeling * * Required update sites: * - IJPB-Plugins (MorpholibJ) */ run("Close All"); // 2D connected components labeling // File › Open... open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary__nuclei.tif"); // Image › Rename... rename("binary"); // 4-connected // Plugins › MorphoLibJ › Binary Images › Connected Components Labeling run("Connected Components Labeling", "connectivity=4 type=[8 bits]"); // Image › Lookup Tables › glasbey_on_dark run("glasbey_on_dark"); // Image › Adjust › Brightness/Contrast... setMinAndMax(0, 20); // 8-connected selectWindow("binary"); run("Connected Components Labeling", "connectivity=8 type=[8 bits]"); run("glasbey_on_dark"); setMinAndMax(0, 20); run("Tile"); // 3D connected components labeling (6-connected) open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xyz_8bit_binary__spots.tif"); rename("binary"); run("Connected Components Labeling", "connectivity=6 type=[8 bits]"); run("glasbey_on_dark"); setMinAndMax(0, 255); // Note: surprisingly this determines the content of below histogram! run("Histogram", "stack");
Exercises
Show exercises for:ImageJ MorpholibJ Macro & GUI
- Open xy_8bit_binary__many_vesicles.tif
- Create a label mask image (beware the output data type, because there are quite many objects).
- Count the number of objects in the label mask image.
Solution
/** * Required update sites: * - IJPB-Plugins (MorpholibJ) */ run("Close All"); // File › Open... open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary__many_vesicles.tif"); run("Connected Components Labeling", "connectivity=4 type=[16 bits]"); run("glasbey_on_dark"); run("Histogram");
Assessment
Fill in the blanks
Fill in the blanks, using these words: less, more, 6, 255, 4, more.
- In 3D, pixels have _____ neighbors than in 2D.
- 8-connected connectivity results in _____ objects than 4-connected connectivity.
- In 3D, pixels have __ non-diagonal neighbors.
- In 2D, pixels have __ non-diagonal neighbors.
- A 8-bit label mask image can have maximally _____ objects.
- The maximum value in a label mask image is equal to or _____ than the number of objects.
Solution
- more
- less
- 6
- 4
- 255
- less
Follow-up material
We recommend reading these modules next:
Learn more: